Here we present ConFiG: Contextual Fibre Growth, a brand-new state-of-the-art numerical phantom generator capable of creating more realistic synthetic white matter tissues than ever before. We do this by mimicking the way axons grow in nature, following chemical cues in their envrionment. The video below shows a ConFiG phantom alongside a series of virtual histological slices, demonstrating the complex morphology ConFiG can generate.
For full details of exactly how we make this happen, check out the full journal article here
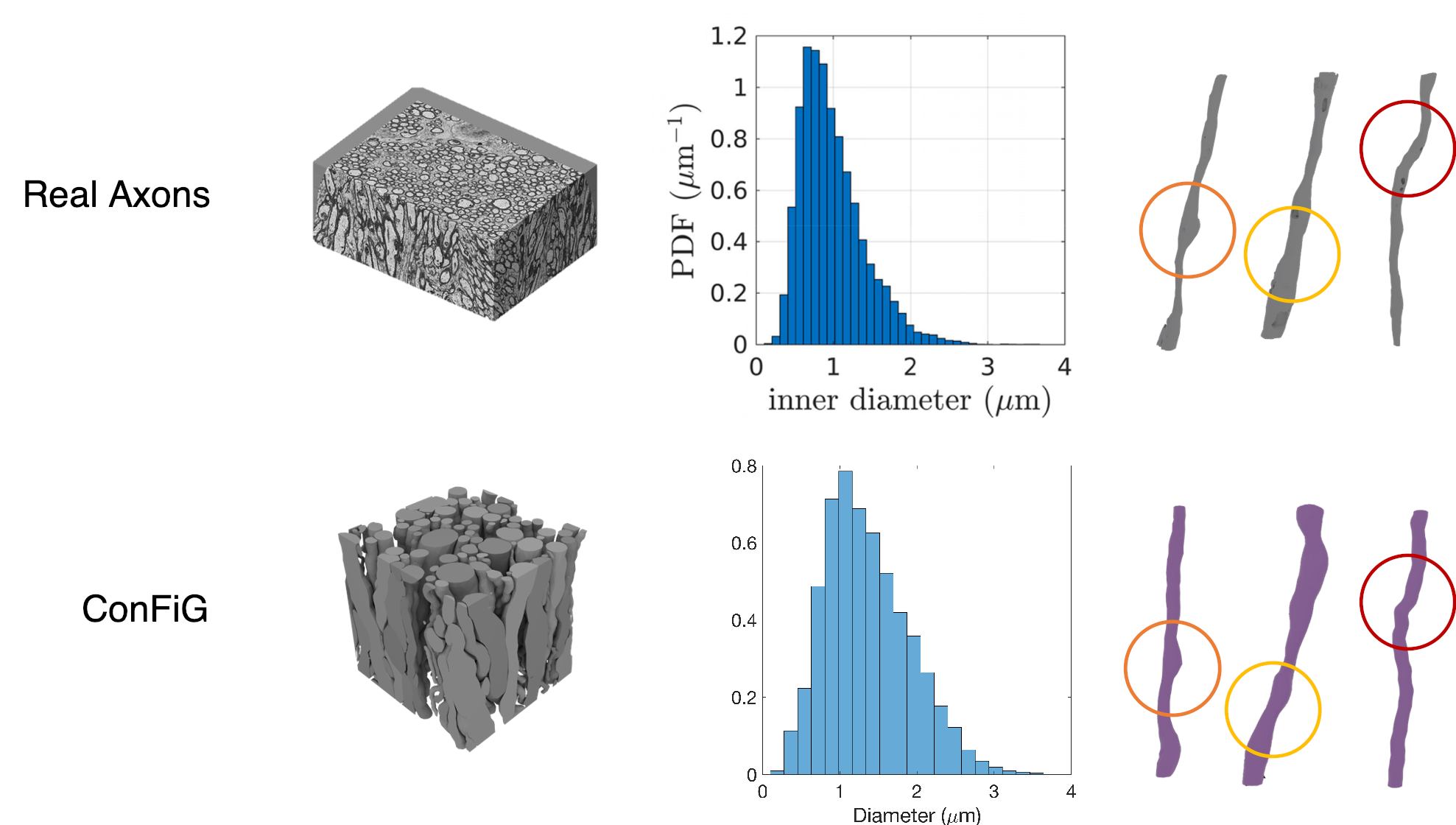
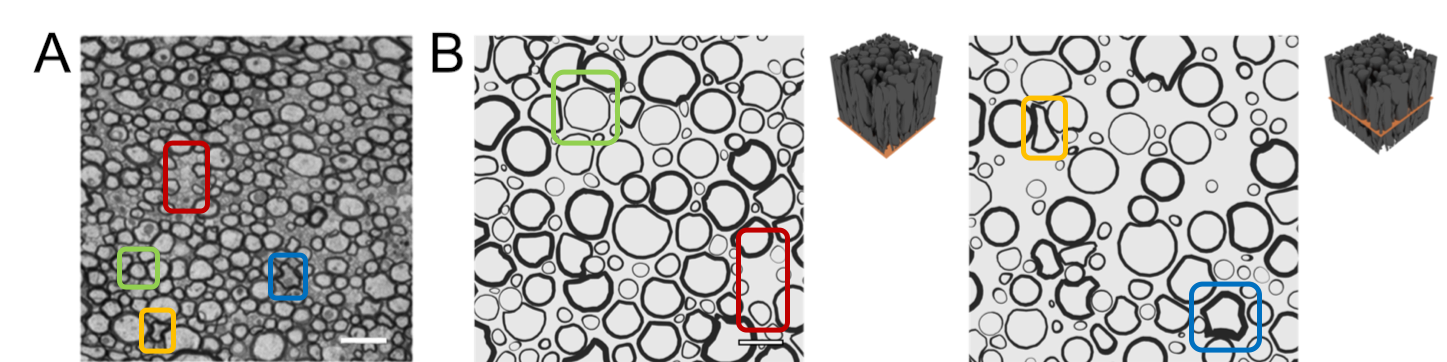
ConFiG generates microstructurally realistic phantoms as evidenced in the below two figures. Firstly, Figure 1 shows that ConFiG captures the diameter profile well when compared to real axons. Not only that, but fine details like the bulges and bends in fibres are seen in both ConFiG and real fibres - note that none of these fine details are explicitly imposed, but just arise organically from the ConFiG growth algorithm.
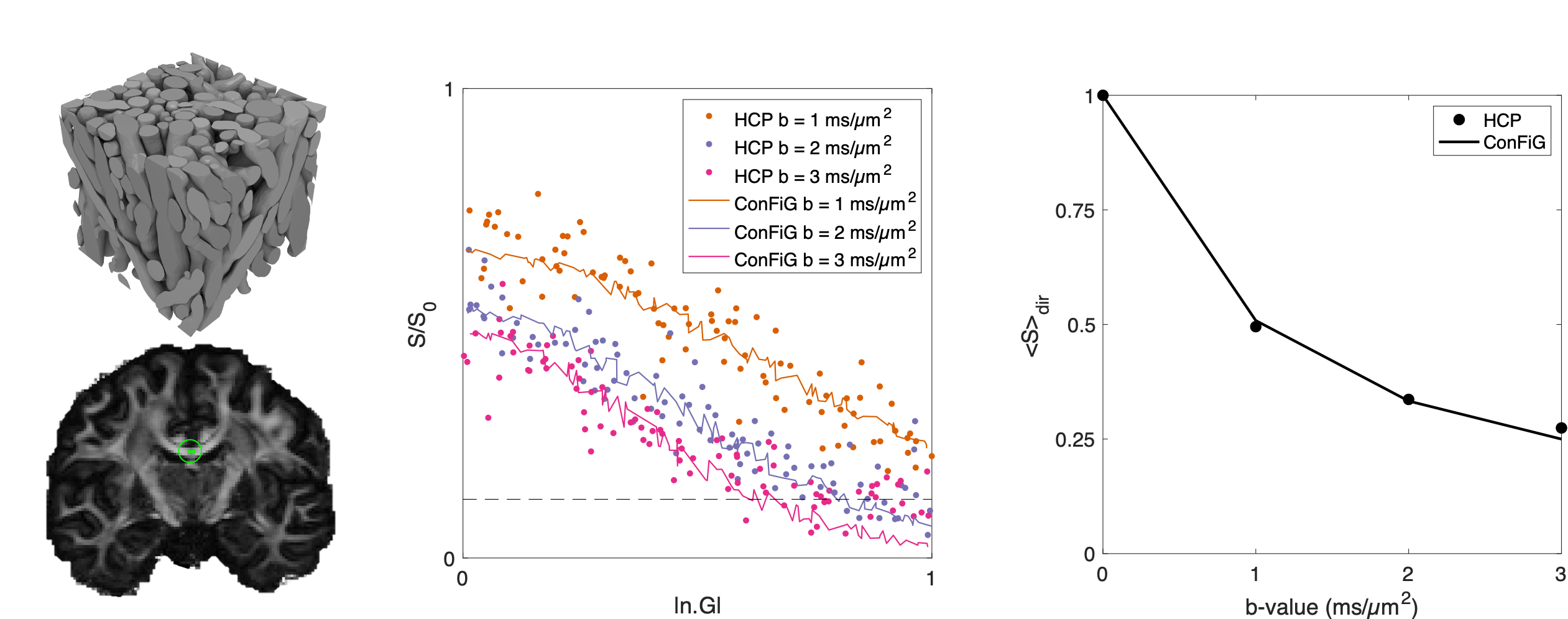
These realistic synthetic tissues also give rise to realistic simulated diffusion MRI (dMRI) signals as shown in Figure 3. Here, we generated three ConFiG phantoms to be representative of three WM regions in the brain. Using these phantoms, we simulated the dMRI signal using the Human Connetome Project (HCP) measurement scheme and compared the simulated signal to real signal from an HCP subject. As Figure 3 shows, the simulated and real signals correspond well, both in terms of direction averaged signal (bottom row) and direction dependent signal (middle row). There is some devation from the real signal at high |n.G| in the middle row (i.e. measuring the signal along the main fibre bundle direction), though this to be expected as the real signal reaches the noise floor and the noise-free simulated signals continue to decay.
ConFiG is a valuable new tool, giving us the capability to investigate microstructural features with more detail than ever before. This opens up avenues for thorough validation of exisiting microstructure imaging methods as well as development of exiting new models for microstructure imaging.